All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme | AYA and adult B-ALL: Two novel high-risk subtypes expressing CDX2 and IDH 1/2 mutations
B-cell acute lymphoblastic leukemia (B-ALL) is a hematologic malignancy that occurs across all age groups and has several classical and genetic subtypes that influence the course of the disease. Adults with B-ALL, for example, tend to have poorer outcomes, and while this has been linked to the prevalence of Philadelphia chromosome (Ph)-negative (Ph−) disease in this population, subtypes of Ph− disease have not been well defined.1
In an attempt to obtain a better understanding of gene expression profiles in adolescents and young adults (AYA), as well as adult patients with Ph− B-ALL, Yasuda et al.1 recently published a study in Blood in which they classified patients into subtypes using combined RNA- and DNA-sequencing analyses. We present a summary of their findings below. Please visit the ALL Hub to see our first article in this educational series here.
Study design
This was a retrospective study of AYA and adult patients (n = 354), aged 15–64 years, with newly diagnosed Ph− B-ALL enrolled in the ALL202-U (n = 54), ALL202-O (n = 147), and Ph− B-ALL213 (n = 153) studies conducted by the Japan Adult Leukemia Study Group (JALSG). Eligible patients had available RNA specimens.
- Ph− B-ALL patients between 15 and 24 years of age and between 25 and 64 years of age were enrolled concurrently in ALL202-U and ALL202-O, respectively, from August 2002 to January 2011.
- Ph− B-ALL patients between 15 and 64 years of age were enrolled in the Ph− B-ALL213 study from July 2013 to October 2016.
Detection of gene rearrangements and gene expression analysis
RNA-sequencing (n = 169), target capture (TC)-RNA-sequencing (n = 31), or both (n = 154) was performed on all patients, and libraries were prepared and subjected to next-generation sequencing (NGS). Gene expression profiles were generated from the RNA-sequencing data, and ultimately, 70 reference sequence entries were used for hierarchical cluster analysis in the classification process.
TC-DNA sequencing and whole exome/genome sequencing
Target enrichment sequencing libraries were prepared with a custom capture panel to detect single nucleotide variants (SNVs), copy number variations (CNVs), structural variations, and tumor clonality. These libraries were then subjected to NGS.
- Only deletions of common regions (IKZF1, PAX5, EBF1, CDKN2A/2B, RB1, ETV6, BTLA, TBL1XR1, ERG, NF1, ATM, IKZF2, TP53, CREBBP, VPREB1, NR3C1, and TCF3) were evaluated for focal CNV assessment.
- Candidate focal CNVs were manually reviewed, filtered, and validated.
- Sequencing libraries for whole-exome sequencing (WES; n = 10) and whole-genome sequencing (WGS; n = 3) were prepared to detect SNVs and structural variations.
Results
Classification of B-ALL subtypes in AYA and adult patients
Gene expression profiles and genetic alterations—including rearrangements, SNVs, and CNVs—were obtained for 354 patients with B-ALL using RNA-sequencing, TC-RNA-sequencing, and TC-DNA-sequencing analyses; several clusters with consistent results between the methods were identified using t-distributed stochastic neighbor embedding (tSNE) and hierarchical clustering. The cohort was grouped into subtypes based on rearrangements, gross chromosomal alterations, or both gene expression profile and genetic alterations (Figure 1)..
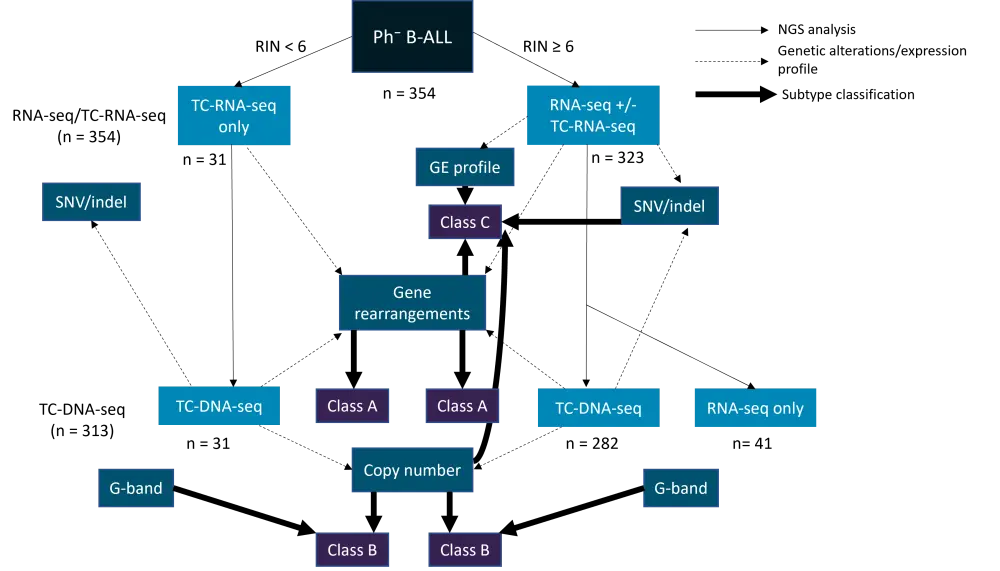
B-ALL, B-cell acute lymphoblastic leukemia; NGS, next-generation sequencing; RIN, RNA integrity number; RNA-seq, RNA sequencing; SNV/indel, single nucleotide variation/insertion or deletion mutation; TC-RNA-seq, target capture RNA sequencing; TC-DNA-seq, target capture DNA sequencing.
Class A: defined by gene rearrangements; Class B: defined by copy number variations; Class C: defined by integration of expression profile and genetic alterations (rearrangements, copy number variations, and mutations).
*Adapted from Yasuda et al.1
RNA sequencing was performed in 323 cases, of which 250 (77.4%) were classified into 14 previously recognized subtypes:
- The most prevalent subtype in AYA (21.6%) and adults (23.8%) was defined by ZNF384 rearrangements.
- Other subtypes defined by rearrangements included:
- TCF3-PBX1 (9.0%)
- DUX4-rearranged (7.4%)
- MEF2D-rearranged (5.3%)
- KMT2A-rearranged (4.3%)
- BCL2/MYC-rearranged (1.9%)
- ETV6-RUNX1 (0.6%)
In addition to the subtypes mentioned above, two novel groups were identified based on specific expression profiles and recurrent genetic abnormalities. One of these groups was characterized by high expression of CDX2, and the other was characterized by IDH1/2 mutations.
Novel subtype with ectopic expression of CDX2
CDX2 is an important regulator of HOX genes during embryonic hematopoiesis, though it is not expressed by normal adult hematopoietic cells; in mouse models, however, ectopic expression of CDX2 transformed hematopoietic stem cells into leukemic cells. In the group identified in this study—the CDX2-high subtype—all members (n = 11) had higher expression of CDX2 compared with other B-ALL subgroups (and compared with normal lymphocytes). High expression of CDX2 protein was confirmed in all tested samples of this subtype.
- Flow cytometry, lineage-specific gene expression profiling, and immunoglobulin rearrangements revealed that the CDX2-high ALL subtype had a B-cell precursor subtype.
- HOX genes were largely repressed in CDX2-high ALL, which suggests that dysregulation of these genes may not be the primary cause of leukemogenesis in this subtype.
- There was upregulation of IGF1R, and enrichment of gene sets associated with the IGF1 pathway.
- TC-DNA-sequencing revealed 1q gain in 7/9 cases of CDX2-high ALL (78%), making 1q gain highly suggestive of this subtype.
- Two patients were not included in the TC-DNA-sequencing analysis; these patients were found to harbor 1q gain via alternative methods.
- Other recurrently mutated genes in this subtype included PAX5 (n = 2), CDKN2A (n = 2), and ATM (n = 2).
Novel subtype with IDH1/2 mutations
IDH1/2 mutations result in the production of 2-HG, an oncometabolite that inhibits TET2 activity, resulting in aberrant methylation. Oncogenic IDH1 R132C (n = 3) and IDH2 R140Q (n = 4) mutations were identified in this analysis, and a cluster enriched with IDH1 (n = 3) or IDH2 (n = 3) mutations was identified via tSNE analysis.
- None of the patients with IDH1/2 mutations had any subtype-defining point lesions, suggesting a subtype driven by IDH1/2 mutation (the IDH1/2-mut subtype).
- Variant allele frequencies of the two identified mutations (IDH1 R132C and IDH2 R140Q) suggest derivation from clonal leukemic cells.
- Flow cytometry, lineage-specific gene expression profiling, and immunoglobulin rearrangements revealed that the IDH1/2-mut ALL subtype had a B-cell precursor subtype.
- Only two genes—NRAS and IKZF1—were recurrently mutated in the IDH1/2-mut subtype, suggesting that impaired expression of certain genes (due to aberrant DNA methylation) may contribute to leukemogenesis.
- Twenty-three candidate tumor-suppressor genes were identified in association with the IDH1/2-mut subtype, including ZEB2 and MEF2C, both of which are integral to the regulation of B-cell development.
Novel subtypes: Clinical characteristics and outcomes
- The median age at diagnosis for the CDX2-high subgroup was 36 years (range, 26‒63), and the median age at diagnosis for the IDH1/2-mut subgroup was 44 years (range, 17‒53).
- When compared with the investigators’ recent study on childhood B-ALL (in which the same DNA analyses were used):
- The frequencies of CDX2-high and IDH1/2-mut subtypes were significantly higher in the AYA/adult cohort than in the childhood cohort, with only three cases of CDX2-high subtype in the childhood cohort, and no cases with hotspot IDH1/2 mutations (both p <0.01).
- Clinical outcomes were widely varied among subtypes:
- CDX2-high, IDH1/2-mut, and Ph-like subtypes were associated with a 5-year overall survival (OS) rate of <40%.
- ZNF384, TCF3-PBX1, DUX4, and MEF2D were associated with a 5-year OS rate of >70%.
- A multivariate model was used to determine whether these subtypes were prognostic factors independent of well-known risk factors such as age and white blood cell (WBC) count:
- TCF3-PBX1 and ZNF384 subtypes were associated with favorable OS independent of age, sex, and WBC count.
- CDX2-high, IDH1/2-mut, and Ph-like subtypes were associated with decreased OS (Table 1).
- WBC counts were considerably lower in the CDX2-high and IDH1/2-mut subtypes compared to other high-risk subtypes (including Ph-like and KMT2A).
Table 1. Risk factors associated with decreased overall survival*
|
CI, confidence interval; HR, hazard ratio; WBC, white blood cell. |
||||||
|
Variables† |
Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
|
HR |
95% CI |
p value |
HR |
95% CI |
p value |
|
|
Subtype |
||||||
|
CDX2-high |
2.86 |
1.29–6.33 |
0.0097 |
3.48 |
1.54‒7.86 |
0.0026 |
|
IDH1/2-mut |
4.68 |
1.67–13.09 |
0.0033 |
4.86 |
1.70‒13.91 |
0.0032 |
|
Ph-like |
2.99 |
1.80–4.97 |
2.3 × 10−5 |
2.04 |
1.16‒3.59 |
0.014 |
|
Age ≥40 years |
2.07 |
1.33–3.23 |
0.0013 |
2.13 |
1.34‒3.38 |
0.0013 |
|
Male sex |
1.29 |
0.83–2.01 |
0.27 |
1.41 |
0.88‒2.26 |
0.15 |
|
WBC ≥30,000/µL |
1.86 |
1.18–2.92 |
0.0074 |
1.81 |
1.07‒3.06 |
0.028 |
Also of note is that high-risk subtypes accounted for 30% of Ph− B-ALL cases in AYA and adults in this study, but accounted for less than 10% of cases in the childhood cohort in the investigators’ previous study.
Screening for CDX2-high and IDH1/2-mut subtypes will help prevent the misidentification of AYA and adult patients who may otherwise be assigned to a lower risk category based on low WBC counts, and can help to ensure that patients receive the correct treatment.
Conclusion
In this genetic analysis of AYA and adult patients with Ph− B-ALL, two novel subtypes were identified: one expressing high amounts of CDX2 and the other characterized by the presence of IDH1/2 mutations. These novel subtypes are associated with poorer OS and may be useful prognostic factors in determining clinical outcome, and they may also—together with other high-risk subtypes—partially account for the difference in prognosis between AYA/adult patients and children. The limitations of this study dictate the need for further investigation, including in vivo functional analyses of CDX2 and IDH1/2 mutations on the development of B-ALL, further clarification of genetic lesions and mechanisms leading to high expression of CDX2, and a larger cohort in which the prognostic impacts can be fully elucidated.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

