All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme | Genetic heterogeneity, incidence, and prognosis of T-ALL
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignant neoplasm accounting for 10–15% of pediatric and 20–25% of adult cases.1 Recent technological advancements have revealed the broad genetic heterogeneity of T-ALL, characterized by the accumulation of several genetic aberrations in key processes for thymocyte development such as cell growth, proliferation, survival, and differentiation; the incidence of these aberrations varies according to age.2 Although the clinical implications of large-scale genetic profiling are currently relatively unknown, there has been some elucidation of their prognostic value for some T-ALL subtypes.3
In this educational theme article, we explore the biology, incidence, and prognostic relevance of T-ALL across all age groups.
Early T-cell precursor ALL and early/late cortical T-ALL
T-ALL can be subclassified into early T-cell precursor (ETP), early cortical, and late cortical based on the unique immunophenotypic and gene expression signatures associated with different stages of arrest within the thymocyte development process (Figure 1).1
The ETP-ALL subtype, representing an early-stage arrest in T-cell development, is more prevalent in adult patients than pediatric patients, representing 40–50% and approximately 10% of cases identified, respectively. This subtype is characterized by lack of expression of the CD4 and CD8 stem cell markers, negative or weak expression of CD5, expression of CD34, CD117, and myeloid markers such as CD13 and CD33. Its genetic signatures include activation of HOXA, MEF2C, or BCL11B,3 mutations in signaling factors (NRAS, FLT3), epigenetic regulators (EZH2, IDH1, IDH2, DNMT3A), transcription factors (RUNX1, GATA3, ETV6), and a lower prevalence of NOTCH1 mutations and CDKN2A deletions (Figure 1). Although ETP-ALL was previously associated with poor outcomes, the introduction of early-response-based intensification therapy has helped overcome this; patients with ETP-ALL can now be successfully treated and achieve favorable outcomes.1
The early cortical T-ALL subtype consists of CD1a+, CD4+, and CD8+ immunophenotypes and activation of the homeobox genes TLX1, TLX3, NKX2.1, and NKX2.2. This subtype is generally characterized by a favorable prognosis. This subtype also presents with the highest expression of NOTCH1 mutations and co-occurring with CDKN2A deletions. In mature cortical T-ALL, immunophenotypic expression includes CD4+, CD8+, CD3+, and activation of TAL/LMO genes (Figure 1).1
Figure 1. T-ALL differentiation stages, alongside their phenotypic and genetic profile*
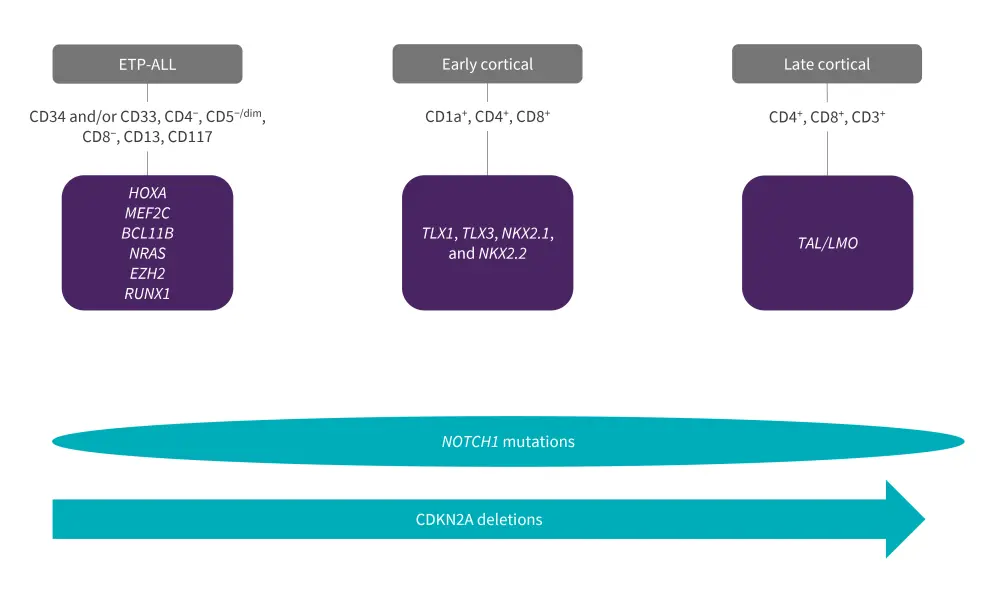
ETP-ALL, early T-cell precursor-acute lymphoblastic leukemia; LMO, LIM domain only; TAL, transcription activator-like.
*Adapted from Gianni, et al.1 and Bardelli, et al.3
Molecular pathways in T-ALL pathogenesis
The pathogenesis of T-ALL involves disruptions in multiple molecular pathways, as seen in Figure 2.4 These include oncogenic NOTCH1 signaling, alterations to cell cycle regulators, increased activation of kinase signaling, transcriptional alterations of oncogenes or tumor-suppressor genes, alterations in ribosomal function and translation, and deregulation of epigenetic regulators.
Figure 2. Molecular pathways involved in T-ALL pathogenesis*
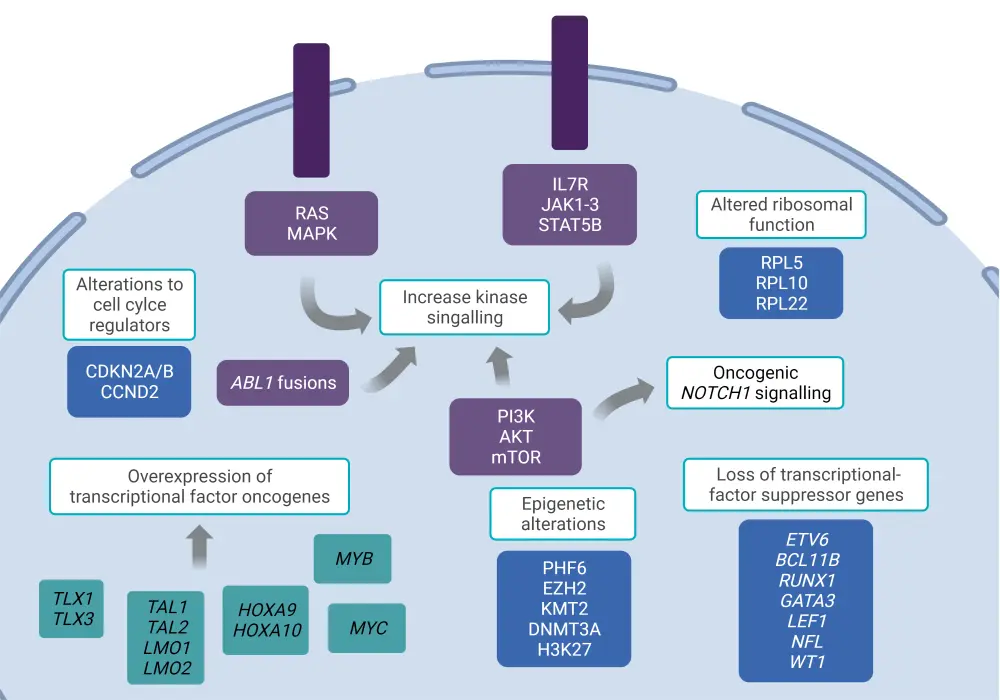
*Adapted from Fattizzo, et al.4 Created with BioRender.com
NOTCH1 signaling
Activation of NOTCH1 signaling is a hallmark event in T-ALL pathogenesis, occurring in more than 60% of cases and affecting T-cell specification and development; NOTCH1 mutations frequently occur alongside CDKN2A deletions. Activated NOTCH1 signaling is generally caused by somatic mutations which influence its function, this occurs through disruption of the negative regulatory region which results in activation independent of ligand binding or via truncation of the PEST domain, impairing the proteasomal degradation of the intracellular domain. Overall, 20% of T-ALL cases present with NOTCH1 negative regulatory region mutations, alongside NOTCH1, PEST, or FBXW7 mutations, which cause aberrant signaling.1
Frequency of NOTCH1 mutations were equally distributed between pediatrics, adolescents and young adults (AYAs), and adults, with an incidence in 61.5%, 60%, and 64% of cases, respectively. A similar pattern was seen for FBXW7 mutations.5
The overall prognostic relevance of NOTCH1 mutations remains unclear, with different studies reporting it as both an unfavorable and favorable prognostic biomarker.3 In pediatric patients, NOTCH1 mutations have been associated with favorable outcomes, with one study reporting higher 5-year event-free survival and overall survival rates among patients with the NOTCH1 mutation with or without FXB73 mutations.6
Cell cycle deregulation
CDKN2A/B deletions, occurring in more than 70% of T-ALL cases, represent another predominant event resulting in abnormal cell cycle control. Aberrations in other cell cycle control proteins, such as the cell cycle regulator RB1 in association with CDKN1B, an inhibitor of cyclin E-CDK2 and cyclin D-CDK4 complexes, are observed in approximately 15% of T-ALL cases.1
Incidence of CDKN2A deletions differed across the three age groups, with significant enrichment in the pediatric cohort (over ~70%), followed by AYAs (~60%) and adults (~42%).5 Owing to the wide heterogeneity of T-ALL and small sample sizes used in pediatric studies, the prognostic value of CDKN2A is currently debated; studies have shown correlations of its deletion with both unfavorable and favorable survival outcomes.6
Transcriptional factors (oncogenes and tumor-suppressor genes)
Overexpression of distinct transcription factor oncogenes such as TAL/LMO, homeobox HOXA, TLX1/3, NKX2-1/2-2, and MEF2C can be classified as type A abnormalities affecting thymocyte development, maturation, and differentiation. Combined, these genetic abnormalities account for up to 70% of T-ALL cases, with varying distribution in pediatrics and adults, as seen in Figure 3.3
Figure 3. Frequency of T-ALL transcriptional factors mutations in adults and children*
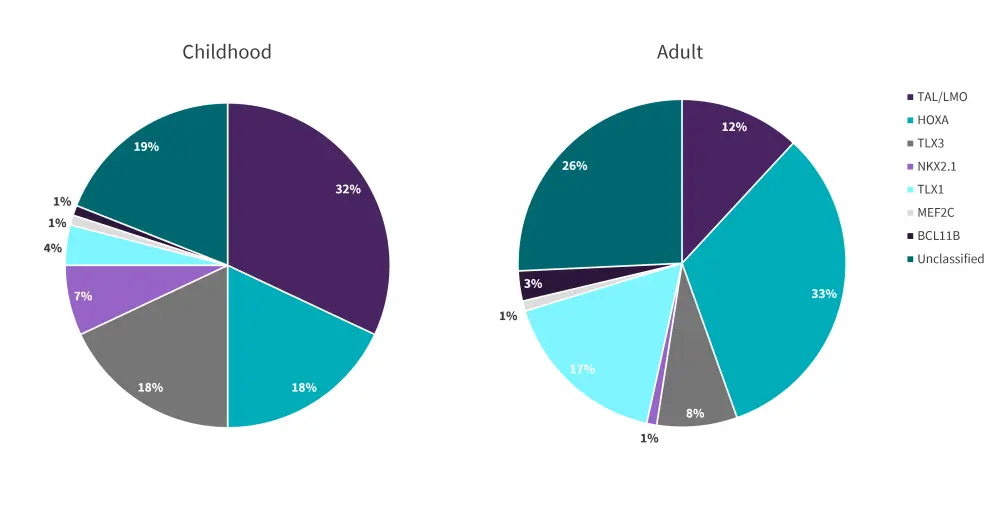
*Adapted from Bardelli, et al.3
TAL and LMO factors share the same transcriptional complex; therefore, their mutations commonly co-occur. This observation suggests a possible collaborative effect in T-ALL pathogenesis which is further influenced by downstream signaling of TRIB2, NKX3.1, RUNX1, GATA3, and MYB. Moreover, the TAL/LMO genes share a similar gene expression signature on T-ALL cells and are often categorized as one subgroup. TAL/LMO mutations are highly expressed in pediatric T-ALL compared with adult T-ALL (Figure 3). Overall, TAL/LMO subgroup mutations are considered prognostic markers of adverse outcomes and have been identified as a poor prognostic marker in children; however, they have conversely shown potential as a predictive biomarker of minimal residual disease.3,6
HOXA-positive T-ALL represents another genetic subgroup of T-ALL, related to a cluster of genes including HOXA, KMT2A, or MLLT10; this specific subtype is more commonly observed in adults compared with children (Figure 3). Unfavorable prognostic outcomes have been consistently shown within several groups of patients with this subtype.3
NK-L homeobox gene rearrangements, which include TLX1, TLX2, NKX2-1, and NKX2-2, are also involved in T-ALL pathogenesis.1 TLX1 aberrations are highly enriched in adult patients compared with pediatric cases. On the other hand, TLX3 gene mutations, which result from the t(5;14)(q35;q32) translocation, are observed at a higher frequency in children compared with adults. NKX2-1- and NKX2-2-rearranged leukemias are more commonly found in childhood T-ALL than adult T-ALL, with an incidence of 7% and <1%, respectively (Figure 3).3
The prognostic value of a positive TLX3 subtype in childhood ALL is unclear, with some studies reporting poor prognosis, good prognosis, and no association.3
Mutations in MEF2C result in the arrest of T-cell differentiation at early stages; therefore, it is commonly found in ETP-ALL. This subgroup is rarely seen in patients, with 1% incidence for both adults and children (Figure 3).3
Other oncogenes deregulated in T-ALL are MYC and MYB. The upregulation of MYC via NOTCH1 signaling contributes to T-cell leukemogenesis.1 Overexpression of MYC via MYC translocations are observed in around 5% of pediatric and adult T-ALL cases, whereas aberrant expression of MYB, due to focal-specific duplications and other mutations, occur in approximately 10% and 19% of T-ALL cases, respectively.1
Deletions or inactivations of tumor-suppressor genes, which mainly involve BCL11B, LEF1, WT1, ETV6, RUNX1, and GATA3, are also linked to T-ALL pathogenesis (Figure 2).1 Deletions of the latter three genes are highly represented in ETP-ALL, while deletions of BCL11B, LEF1, and WT1 are mainly identified in early cortical T-ALL. The common combination of BCL11B alongside TLX1 and TLX3 translocations are seen in up to 10% of T-ALL cases. Considering age distribution, the BCL11B mutation is identified in less than 1% of pediatric and 3% of adult cases (Figure 3). LEF1 and WT1 mutations are seen in 15% and 10%, respectively, for both pediatric and adult T-ALL.1
Of ETV6, RUNX1, and GATA3 deletions, ETV6 has the highest incidence of 13%, with a lower prevalence seen for the RUNX1 and GATA3 subtypes.1 Overall, the prognostic outlook is poor for these subtypes.4
Signaling pathways
Increased kinase signaling due to mutations in the components of the IL7R/JAK/STAT, PI3K/AKT/mTOR, RAS/MAPK, and ABL kinase pathways, are involved in T-ALL pathogenesis (Figure 3).3
The most common alteration involved in constitutive activation of the PI3K/AKT/mTOR pathway is the loss of function/inactivation of the tumor-suppressor gene PTEN, which is seen in 10–15% of cases overall. Less frequently observed mutations within oncogenic signaling of the PI3K/AKT/mTOR pathway, such as activating mutations involving AKT1 and the PI3K catalytic and regulatory subunit genes PIKC3A and PIK3CD, have also been shown to play a role in T-cell pathogenesis.1
The prognostic effect of PTEN mutations has not been fully established; however, PTEN deletions have shown potential as a predictive biomarker of lower event-free survival and overall survival outcomes. Also, many genes involved in hyperactivation of the PI3K/AKT/mTOR pathway are predictive of glucocorticoid resistance in T-ALL.3 Observed in up to 27% of pediatric cases, PTEN deletions were found to be associated with unfavorable treatment and survival outcomes.6
The IL7-R mediated JAK-STAT pathway plays a key role in the proliferation and survival of T-cell development in the early stages, hence its predominant association with the ETP-ALL subtype when dysregulated. Its overactivation is driven by several mutations, including IL7R, JAK1, JAK3, and STAT5, which accounts for 25% of T-ALL cases.1
Individually, IL7R, JAK1, JAK3, and STATB5 mutations are identified in 10%, 7%, 4%, and 1% of T-ALL cases, respectively.3
Hyperactivation of the RAS/MAPK pathway, due to mutations in HRAS and KRAS oncogenes, is involved in 5–10% of T-ALL cases. Other deregulated components of this pathway include mutations in NF1 and PTPN11, observed in up to 3% of cases.4 In pediatric T-ALL, genomic abnormalities that result in overactivation of the RAS/MAPK pathway are particularly prevalent at relapse (40% of cases), while their incidence at diagnosis is much lower (15%).3 Among this subset of patients, RAS mutations have been characterized as a high-risk feature, associated with poor clinical outcomes and resistance to steroid treatments.6
The NUP214-ABL1 fusion is the most common genetic abnormality of the ABL gene, detected in 6% of pediatric T-ALL cases. This genetic alteration has been linked to high-risk relapse and treatment failure in childhood T-ALL; however, some rearrangements have shown sensitivity to TKI inhibitors.6
Frequent genetic alterations of epigenetic regulators in T-ALL include the following: PHF6, KDM6A, EED, EZH2, and SUZ12. Deletions or loss-of-function mutations in PHF6 occur at a higher frequency in adults compared with children (30–40% and 15%, respectively).3,6 EZH2, EED, and SUZ12, encompassing the polycomb repressive complex 2 (PRC2) and involved in the methylation of H3K27, are mutated in about 25–30% of T-ALL cases and have been associated with poor responses to chemotherapy. Loss-of-function mutation of KDM6A is seen in approximately 5–15% of T-ALLs.3
Mutations in ribosomal genes involved in cell proliferation, namely RPL5 (1p22), RPL10 (Xq28), and RPL1, have been identified in T-ALL genetic profiling studies. Mutations of RPL10, most of which affect the RPL10 R98S allele, have been identified in 5–10% of pediatric T-ALL cases and have been associated with mutated Bcl2.1,6
Conclusion
Large-scale genetic profiling has helped identify relevant genetic alteration patterns in T-ALL, evaluate the association between genetic subtypes, and classify genetic subtypes within thymocyte development. Given the wide genetic heterogeneity of T-ALL, identifying the specific prognostic relevance for all existing genetic subtypes remains a challenge. More efforts are needed to help translate the complex biology of T-ALL in the clinical setting.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

