All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme | The role of WGS and WTS for the genetic classification of adult ALL
The focus of our educational theme this quarter on the ALL hub is the role of sequencing in ALL disease management. In this first article of the series, the focus will be on the role of whole genome sequencing (WGS) and whole transcriptome sequencing (WTS) for diagnostic classification.
Molecular subtypes are established predictors of clinical outcome in children with acute lymphoblastic leukemia (ALL) and are essential components for risk stratification. Historically, adult B-cell ALL (B-ALL) has been underrepresented in large genomic analyses, although more recent work has demonstrated that molecular subtypes could have a therapeutic role in this age group as well. Whilst genomic information is clearly important, the heterogeneity of B-ALL means that a number of different assays are currently required to make an accurate molecular diagnosis.
The application of World Health Organization (WHO) classification and European Leukemia Network guidelines require a combination of cytogenetics and targeted sequencing to detect specific mutations needed for diagnosis, prognostication, and therapy selection. The current standard-of care (SoC) techniques include chromosome banding analysis, fluorescence in situ hybridization (FISH), and panel sequencing to identify structural variants (SV), copy number variations, and mutations. These tests are often performed sequentially due to the associated costs and complexity and may still fail to identify some newer groups which are defined by a combination of genomic features rather than a single event. More recently developed techniques like optical genome mapping and whole exome sequencing, can also identify a subset of these genetic abnormalities; however, only WGS is capable of assessing all subtypes of genetic abnormalities. WTS is another reliable single assay that allows the simultaneous, comprehensive, analysis and identification of all transcriptional events.
Herein, we provide a summary of recent data from abstracts presented at the 63rd American Society of Hematology (ASH) Annual Meeting and Exposition regarding the diagnostic utility of WGS and WTS for the genetic characterization of adult ALL.
The role of WGS and WTS in diagnostic classification
Haferlach talked about WGS and WTS as diagnostic tools for the genetic characterization ALL in adults.1 The study cohort comprised of 293 patients with B-ALL and 124 with T-cell ALL (T-ALL) that had been diagnosed in a routine setting with current SoC techniques. These cases were also evaluated by WGS with a median coverage of 90×. Different tools were used to evaluate copy number variations, copy neutral loss of heterozygosity, SV, small nucleotide variants, and indels. Further analysis with WTS was used to determine transcript and gene expression profiles as well as fusions.
Ryland also presented results on the diagnostic utility of multimodal genomic profiling for the molecular classification of adult B-ALL.2 The study aimed to assess the feasibility of comprehensive profiling in adult B-ALL by performing prospective WGS and WTS. The clinical utility of this upfront approach for making a molecular subtype classification was compared to SoC diagnostics (conventional G-ban chromosome analysis, BCR-ABL1 and KMT2A FISH). The secondary aim was to explore the diagnostic uses of this data. From February 2019 to June 2021, 42 consecutive patients with B-ALL, with a median age of 43 years, were recruited. In total, 80% of the cohort were tested at diagnosis and 20 patients met the criteria for a WHO entity based on a recurrent genetic abnormality, which was detected by conventional cytogenetics or FISH, with the remainder having an abnormal but not specific (n = 10) or normal (n = 11) karyotype.
WGS and WTS vs SoC in B-ALL1,2
In both studies, WGS and WTS improved B-ALL subtype classification compared to SoC (Figure 1 and 2).
In the study by Ryland, et al.,2 (Figure 1) six cases remained unclassified, of which two cases were noted as having high CDX2 expression and clustering with other CDX2-expressing B-ALL. These cases likely represent examples of the recently described CDX2 high subtype. Including these two cases in the ALL classification rate for WGS would increase the classification rate to 91%.
Two cases were predicted to be Philadelphia (Ph)-like by ALLSorts, a hierarchical WTS-based classifier, but a genomic lesion could not be identified so they were conservatively classified as B-other. Novel immunoglobulin heavy chain translocations were also common in the B-other group but without a common partner.
Figure 1. WGS/WTS vs SoC testing for B-ALL subtype classification*
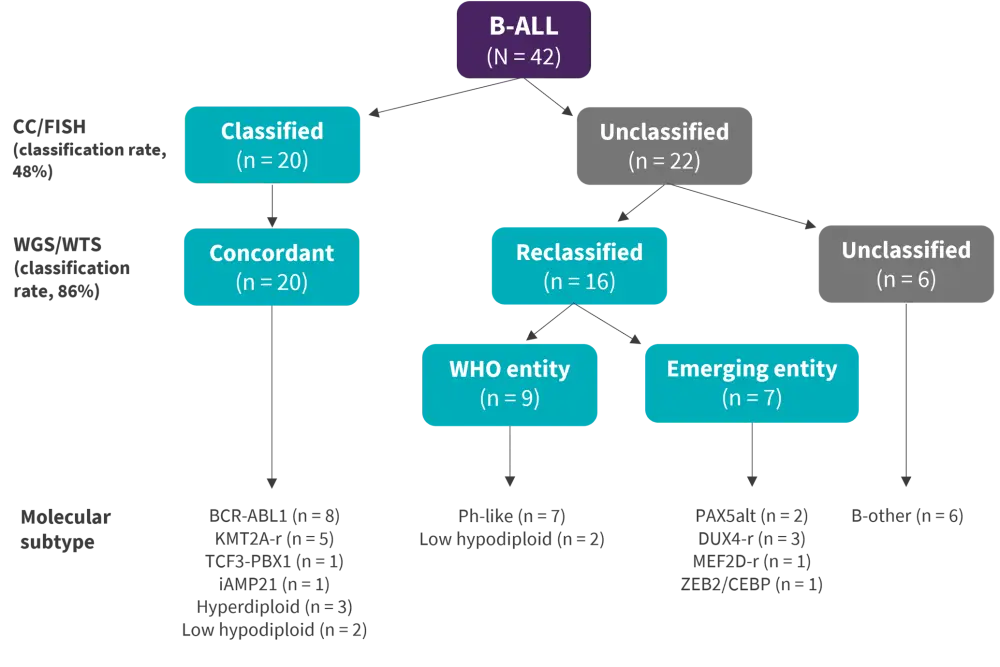
ALL, acute lymphoblastic leukemia; CC, conventional cytogenetics; FISH, fluorescence in situ hybridization; Ph, Philadelphia; r, rearrangement; WGS, whole genome sequencing; WHO, World Health Organization; WTS, whole transcriptome sequencing.
*Adapted from Ryland, et al.2
In the study by Haferlach, et al.,1 to assess discrepancies between the methods, B-ALL cases were categorized into the WHO genetic classes using the SoC techniques and were subsequently reclassified with WGS and WTS. All cases that were assigned to a distinct genetic category based on the SoC techniques were classified in the same genetic category by WGS and WTS. Based on the SoC techniques 110 cases could not be assigned to any of the genetic categories (“other” B-ALL) (Figure 2).
- WGS and WTS identified six cases of low hyperdiploidy that were missed by the SoC techniques due to insufficient proliferation in vitro.
- Eight additional cases of the BCR-ABL1-like category were identified.
- In total, 28 cases were assigned into the novel B-ALL subtypes DUX4-rearranged/ERG deregulated and ZNF384-rearranged.
Figure 2. SoC vs WGS and WTS in “other” B-ALL genetic classification*
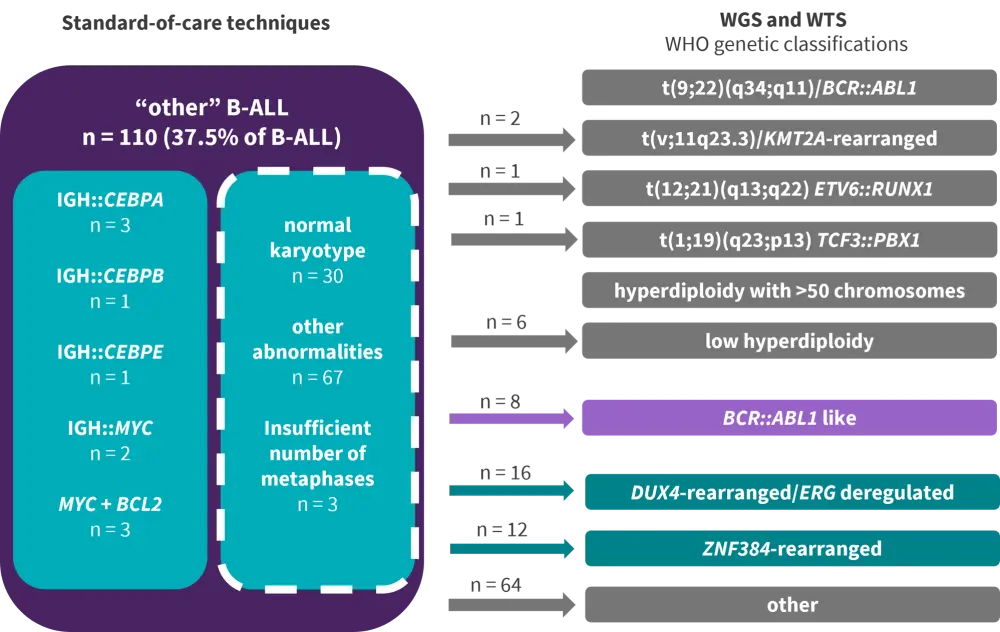
ALL, acute lymphoblastic leukemia; WGS, whole genome sequencing; WHO, World Health Organization; WTS, whole transcriptome sequencing
*Adapted from Haferlach, et al.1
About the novel B-ALL subtypes
BCR-ABL1-like B-ALL:
- Lacks the BCR-ABL1 rearrangement but has a similar gene expression profile to BCR-ABL1 ALL, with translocations involving tyrosine kinases or rearrangements involving CRLF2 or the EPOR receptor.
- Of all B-ALL cases, 41 cases (14%) were assigned by gene expression profiling to the BCR-ABL1-like group, while 44 cases were identified by combined WTS and WGS fusion analysis. There was an overlap of 38 cases between the two methods.1
DUX4-rearranged B-ALL:
- cannot be identified based on chromosome banding analysis analysis or FISH due to the small size of the insertions.
- Difficult to identify by WGS due to the complexity of the genomic region and the presence of multiple DUX4 copies in the genome; the only technique so far which is able to identify this subset of ALL is gene expression profiling.
- Has a more favorable prognosis than the “other” B-ALLs that are not characterized by a specific genetic abnormality.
ZNF384-rearranged B-ALL:
- Difficult to detect in routine diagnostic because the most frequent rearrangement is cytogenetically cryptic, but it can be easily identified by WGS.
- Has a more favorable prognosis than the “other” B-ALLs that are not characterized by a specific genetic abnormality.
SoC vs WGS and WTS in T-ALL1
In the study by Haferlach, et al., T-cell ALL (T-ALL) cases were assigned according to the SoC into aberrant (65%), normal (28%), and non-evaluable (7%) karyotype. The subgroups were then revaluated using WGS and WTS. Using these methods all T-ALL cases were able to be characterized.
Non-evaluable cases by SoC showed 100% aberrant karyotype with WGS and WTS. The normal karyotype cases based on SoC were found to be 80% aberrant and 20% normal. Typical T-ALL abnormalities where aberrant by both SoC techniques and WGS and WTS (100%). Overall, using WGS and WTS, abnormalities were found in 94.4% of all T-ALL cases, while 5.6% were characterized as normal.
WGS vs WTS2
In the Ryland study, the contribution of genome vs transcriptome in making a molecular diagnosis was assessed in the 36 patients that had a true classification.
- Combined WGS-WTS resolved more cases than WGS-only or WTS-only.
- Genome and transcriptome data resulted in an equivalent classification for 22 cases (60%), namely, a (9;22) translocation at the genome level, a BCR-ABL1 fusion by WTS, a Ph ALLSorts prediction, a typical pattern of chromosome loss, TP53 mutation at the genome level, and a low-hypodiploid ALLSorts prediction.
- WGS was more informative in two cases. One of these cases was iAMP21, which was clearly identified by WGS, but with an incorrect ALLSorts classification, highlighting the difficulty in finding a specific expression signature for some of the aneuploid subtypes.
- One case was classified as ZEB2 by genome. This subtype was not included in the classifier as there were insufficient numbers of known cases to allow for inclusion in the training set.
- ALLSorts failed to classify five cases with uncommon KMT2A rearrangement partner, although the fusion was identified by WTS and the translocation by WGS.
- Conversely, WTS was more informative in three cases, which were predicted to be DUX4 by ALLSorts. In all three the SV could be identified by WGS, but only upon guided reanalysis. This is a known challenge with DUX4 rearrangements of the genome level given the multiple copies and genomic location.
- Importantly, both genome and transcriptome analysis contributed to the classification of nine cases. These were all Ph-like or PAX5alt and required the expression profile in combination with the genomic lesions, which is characteristic of these subtypes.
Conclusion
In conclusion, WGS and WTS were able to reliably detect the abnormalities that were identified by SoC techniques. WGS and WTS could comprehensively characterize the whole genome with a higher resolution compared to the current SoC techniques, providing additional relevant information for prognostication and therapy selection in adult B-ALL that extends beyond WHO entities. The turnaround time of 5 to 7 days already meets the diagnostic requirements and both technologies can be implemented with automated analysis pipelines, thus reducing time and error rates. Furthermore, the combination of WGS and WTS may be superior to either alone, especially in the diagnostic setting, where a high level of certainty is required. However, a small proportion of B-ALL will remain unclassified following comprehensive genomics.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

