All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
HLA (mis)matching in haploidentical transplantation
To reduce the risk of graft-versus-host disease (GvHD) 8/8 human leukocyte antigen (HLA) matching is preferred, however the chance of finding a perfect match is low especially for some ethnic groups. By using haploidentical (haplo) transplants, the donor pool is greatly expanded and may allow much more rapid transplantation to be performed.
The use of post-transplant cyclophosphamide (PTCy) after transplant has dramatically reduced the incidence of acute and chronic GvHD. However, the ideal pattern of HLA (mis)matching is unclear and was investigated in a paper by Fuchs et al.1
Study design
Data from the Center for International Blood and Marrow Transplant Research (CIBMTR) of 1,434 patients who underwent a haplo transplant and the patient characteristics are shown in Table 1. Patient characteristics were well balanced between the different subgroups. In the whole study men were more likely to be included with a median age of 54 years. The majority (58%) of patients were diagnosed with acute myeloid leukemia (AML).
Table 1. Baseline patient characteristics1
|
*Including intermediate – cytogenetics not collected. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; HLA, human leukocyte antigen; MDS, myelodysplastic syndrome |
|||||
|
Characteristic |
Total (N = 1434) |
HLA-B leader matched (n = 878) |
HLA-B-leader mismatched (n = 556) |
HLA-DRB1 matched (n = 217) |
HLA-DRB1 mismatched (n = 1215) |
|---|---|---|---|---|---|
|
Recipient age at transplant |
|
|
|
|
|
|
Median (range) |
54 (1−78) |
55 (1-78) |
53 (1-78) |
57 (1-78) |
53 (1-78) |
|
Sex (%) |
|
|
|
|
|
|
Female |
40 |
40 |
40 |
39 |
40 |
|
Disease (%) |
|
|
|
|
|
|
AML |
58 |
58 |
58 |
63 |
57 |
|
ALL |
23 |
23 |
23 |
19 |
24 |
|
MDS |
19 |
19 |
19 |
18 |
19 |
|
Disease risk |
|
|
|
|
|
|
Low |
4 |
4 |
4 |
2 |
4 |
|
Intermediate* |
51 |
52 |
50 |
53 |
50 |
|
High |
26 |
27 |
25 |
27 |
26 |
|
Very High |
3 |
3 |
3 |
2 |
3 |
|
Missing or N/A |
16 |
15 |
19 |
15 |
17 |
|
Ethnicity (%) |
|
|
|
|
|
|
Caucasian-non-Hispanic |
57 |
57 |
56 |
58 |
56 |
|
African American |
16 |
15 |
17 |
20 |
15 |
|
Asian |
6 |
7 |
5 |
7 |
6 |
|
Hispanic |
15 |
14 |
15 |
9 |
16 |
|
Other |
1 |
1 |
1 |
1 |
1 |
|
Graft type |
|
|
|
|
|
|
Bone marrow |
43 |
44 |
41 |
38 |
44 |
|
Peripheral blood |
57 |
56 |
59 |
62 |
56 |
Results
GvHD and survival outcomes
For the entire cohort the Day 100 cumulative incidence of acute GvHD was:
- Grades II-IV 34.7%
- Grades III-IV 9.3%
Chronic GvHD at 1-year was 25.8%.
The three-year cumulative incidence of:
- Relapse 39.1%
- NRM 19.7%
Three-year overall survival (OS) was 52.5% while disease free survival (DFS) was 41.3%.
HLA mismatching and association with clinical outcome
Most of the patients in the study were 5/10 (65%) or 6/10 (22%) matched for HLA-A, B, C, DRB1 and DQB1.
The association between number of mismatches and outcome was assessed. Patients with 5/10 matches compared with patients with 6/10 matched showed a hazard ratio (HR) for relapse of 1.06 (95% CI, 0.87−1.28). Compared with patients 5/10 matched, the HR was 1.35 (95% CI, 1.08 - 1.69) for ≥7/10 HLA matched.
The risk of GvHD, DFS, transplant related mortality or overall mortality were not significantly associated with the total number of matched alleles.
Locus specific risks
The clinical outcome of matching at HLA-A, B, C, DRB1 and DQB1 individually, was assessed. HLA-B was examined in terms of classical matching and leader matching. No significant association was seen with any endpoint for mismatches at HLA-A,-B or -DQB1 alone.
HLA-B leader mismatching however was associated with a significantly decreased OS (HR, 1.23; 95% CI, 1.07 - 2.18; p = 0.004) and increase transplant-related mortality (TRM; HR, 1.43; 95% CI, 1.13 - 1.82; p = 0.003) compared with leader matching. Comparing HLA-B matched (whole sequence matched including leader) with leader matched there was not a significant increase in mortality or TRM, indicating HLA-B leader matching is more clinically relevant.
A significantly reduced risk of relapse was found for HLA-DRB1 mismatching in the GvHD direction versus matching (HR, 0.65; 95% CI, 0.53 to 0.80; p < 0.0001).
Relative to HLA-DRB1-T-cell epitopes (TCE)-permissive matches, mismatches were associated with significantly increased OS s (HR, 0.59; 95% CI, 0.43 - 0.82; p = 0.002).
The risk of acute GvHD was not associated with (mis)matching at any locus. However, the risk of chronic GvHD was increased when HLA-C was mismatched compared to matched (HR, 1.46; 95% CI, 1.17 - 1.83, p = 0.0008).
Mismatching at 2-loci
HLA-B leader mismatching
HLA-B leader mismatching was examined with HLA-DRB1. HLA-B leader matching and HLA-DRB1 mismatching was additive for patients with this combination who comprised >1/2 of the group (Figure 1) and had improved DFS and a reduced relapse rate compared to patients with HLA-B leader mismatch/HLA-DRB1 match status.
Figure 1. HLA-B leader and HLA-DRB1 (mis)matching1
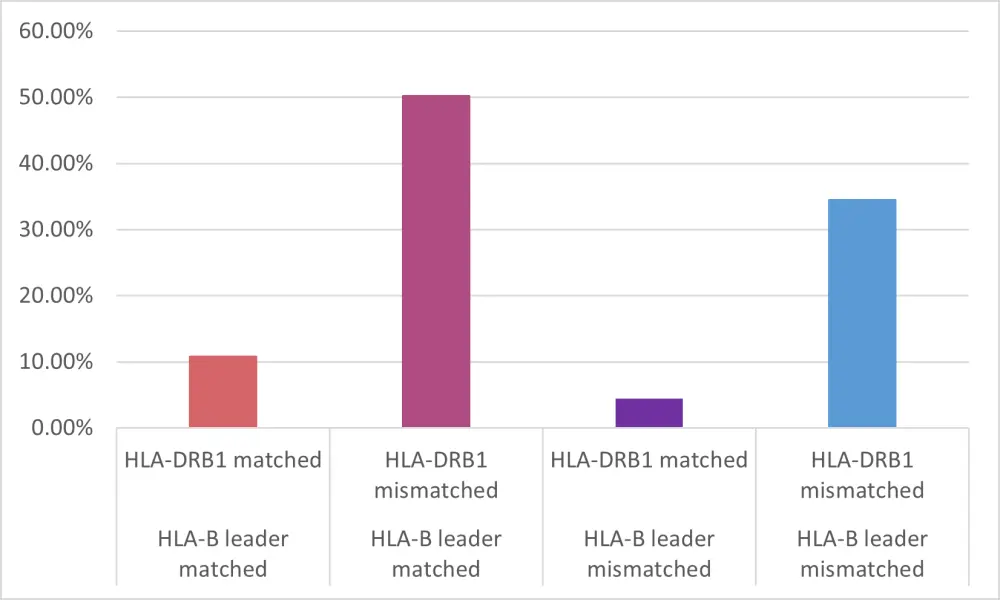
HLA, human leukocyte antigen
HLA-B leader matching/ HLA-DPB1 non-permissive mismatching compared to any other combination of (mis)matching at these two alleles showed longest survival.
HLA-DRB1 and -DQB1 mismatching
Mismatching at both HLA-DRB1 and -DQB1 was favored due to linkage disequilibrium with a mismatch at only one locus occurring less often. The proportion of matched/ mismatched alleles are shown in Figure 2 and are consistent with the expected genetic relationship. The longest DFS was seen in patients with HLA-DRB1-mismatched/DQB1-matched status.
Figure 1. HLA-DQB1 and HLA-DRB1 (mis)matching1
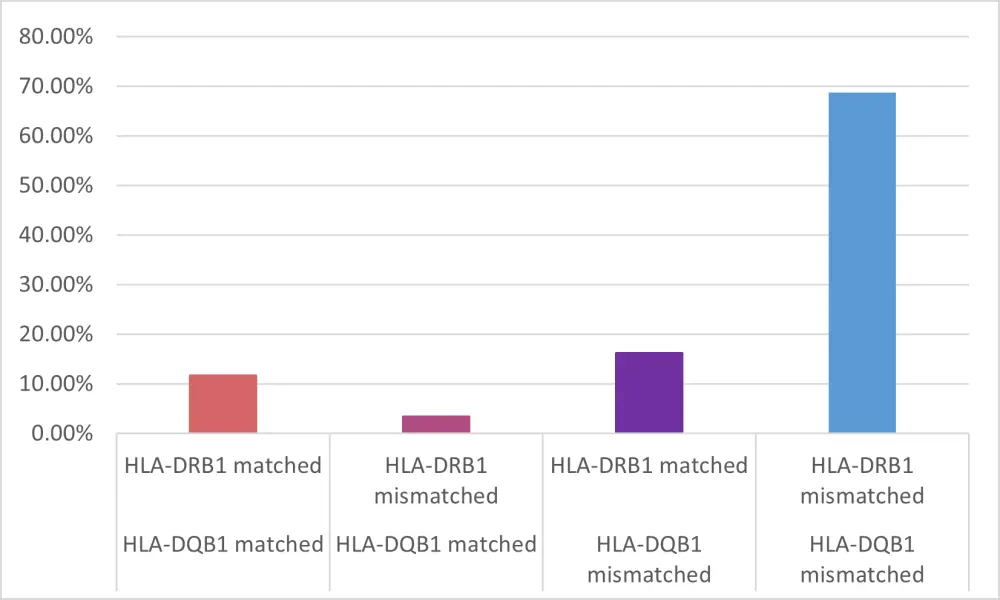
HLA, human leukocyte antigen
Optimal donor selection
From this study the optimal haploidentical donor with respect to improved DFS was found to be:
- HLA-B leader-matched,
- HLA-DRB1-mismatched,
- HLA-DQB1-matched, and
- HLA-DPB1 TCE-non-permissive mismatched.
For patients who are cytomegalovirus negative, a negative donor is preferred.
Conclusion
Using haplo-donors increases the speed at which patients can receive a transplant as most patients may have more than one haplo donor available to them. A novel association of HLA-C and B with survival outcomes following haplo-HSCT + PTCy was identified. For patients treated with PTCy following a haplo-HSCT the ideal donor was HLA-B leader-matched, HLA-DRB1-mismatched and HLA-DQB1-matched, and nonpermissive TCE HLA-DPB1 mismatched.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

