All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Using whole transcriptome sequencing for the genetic stratification of patients with ALL
The 2017 revision to the World Health Organization (WHO) classification of acute lymphoblastic leukemia (ALL) recognizes nine different sub-entities within the B-cell lineage ALL (B-ALL) with recurrent genetic abnormalities, including groups defined by specific translocations, somatic mutations and genes associated with predisposition to neoplasms. The genetic heterogeneity of ALL calls for a single reliable assay that allows the simultaneous, comprehensive analysis, and identification of all transcriptional events. However, the current diagnostic methods require various analyses based on chromosomal banding analysis (CBA) and fluorescence in situ hybridization, which are limited to only classifying patients with well established abnormalities.
In a recently published study by Walter, et al. the diagnostic potential of whole transcriptome sequencing (WTS) for the precise genetic characterization of ALL and its applicability in clinical practice was assessed.1 Key findings are summarized herein.
Study design
In total, 279 patients were selected for whole genome sequencing (WGS) and WTS and compared to a control group with 64 healthy individuals; their characteristics are illustrated in Table 1.
Table 1. WTS analysis patient characteristics*
|
ALL, acute lymphoblastic leukemia; B-ALL, B-cell ALL; T-ALL, T-cell ALL. |
||
|
Characteristic |
Study arm (n = 279) |
Control arm (n = 64) |
|---|---|---|
|
Female/Male ratio, % |
41/59 |
45/55 |
|
Median age (range), years |
49 (0.1–93) |
— |
|
B-ALL |
N = 211 |
— |
|
T-ALL |
N = 68 |
— |
- CBA and fluorescence in situ hybridization were performed in all 279 patients according to the International System for Human Cytogenomic Nomenclature 2016 guidelines and array-CGH analyses were carried out for 123 patients.
- DNA and RNA-based genotyping was used to ensure correct WTS-WGS pairing and gene expression analysis was used to distinguish between B or T lineage.
- Subclassification of B-ALL samples was done progressively, assessing first the presence of chromosomal rearrangements by the means of fusion calling on WTS data.
- Structural variants and somatic copy number variants (CNV) were detected on WGS data and specific gene deletions (IKZF1, CDKN2A, RB1) were identified by matching structural variants and CNV.
- CNV were inferred from raw gene counts to identify relevant ploidy groups, patients were categorized as high hyperdiploid if at least 2 of the following chromosome number abnormalities were gained: 4, 6, 10, 14, 17, 18, and 21.
- The WTS data was analyzed for selected small nucleotide variants in CRLF2, DUX4, JAK2, KRAS, NRAS, PAX5 and TP53.
- Gene expression profiling was used to identify BCR-ABL1-like signatures.
Results
Single nucleotide polymorphisms (SNP) profiles verified correct WGS/WTS pairing. In 278/279 patients, the best matching WGS sample belonged to the same patient as the WTS sample (minimal allele concordance, 0.81). Gene expression analysis reliably assigned samples to the B-ALL or T-ALL group (Figure 1).
Figure 1. Classification tree*
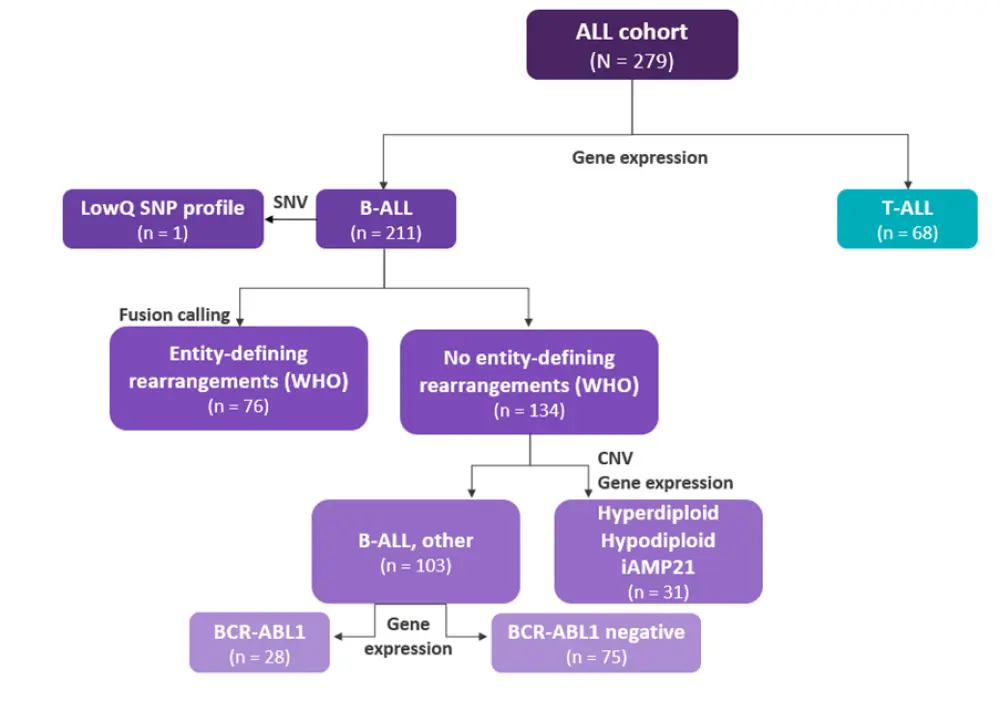
ALL, acute lymphoblastic leukemia; B-ALL, B-cell ALL; CNV, copy number variation; iAMP21, intrachromosomal amplification of chromosome 21; LowQ, low quality; SNP, single nucleotide polymorphism; SNV, structural nucleotide variant; T-ALL, T-cell ALL; WHO, World Health Organization.
*Adapted from Walter, et al.1
Fusion calling
Compared to the standard method of CBA, 97% of the recurrent risk-stratifying gene fusions could be identified by WTS (Table 2), and 76/211 B-ALL patients could be assigned to their respective subtypes. The median number of fusions per patient was 1 (range, 0–8).
Table 2. Comparison of detected fusion transcripts by RNA sequencing to expected translocations by CBA*
|
CBA, chromosome binding analysis. |
|||
|
Fusion transcript |
CBA, n |
RNA sequencing, n |
RNA sequencing/CBA, % |
|---|---|---|---|
|
BCR-ABL1 |
41 |
40 |
98 |
|
ETV6-RUNX1 |
5 |
4 |
80 |
|
KMT2A-AFF1 |
23 |
23 |
100 |
|
TCF3-PBX1 |
4 |
4 |
100 |
In addition to the subtype defining rearrangements, other fusions in the B-ALL cohort were detected. These included read-through fusions such as eight ZNF384 and three PAX5, as well as two fusion transcripts containing NUTM1: CHD4-NUMT1 (a novel fusion) and BRD9-NUTM1 (previously identified in infants with ALL). ZNF384 has been recently described as a new molecular B-ALL ‘other’ subtype with a good response to treatment with prednisone and conventional chemotherapy.
A total of 57 novel fusion transcripts with yet undetermined diagnostic value were also identified by WTS, including genes implicated in other non-hematologic malignancies.
From the remaining 134 patients with B-ALL who had no subtype defining rearrangements, 17 (94%) patient samples were identified as low hypodiploid/near-triploid ALL and 12 (80%) as high hyperdiploid ALL.
BCR-ABL1 expression
Patients with BCR-ABL1-like ALL typically have poor outcomes on conventional treatment strategies; however, there is a potential benefit of treatment with tyrosine kinase inhibitors for this newly defined subgroup. Gene expression profiling detected a BCR-ABL1-like signature in 27% of the remaining patient samples (n = 28), their key characteristics are summarized in Table 3.
Table 3. Characteristics of BCR-ABL1-like ALL vs non BCR-ABL1-like ALL*
|
*Data from Walter, et al.1 |
|||
|
Characteristic |
BCR-ABL1-like |
Non BCR-ABL1-like |
p value |
|---|---|---|---|
|
Elevated white blood cell counts, ×109/L |
59.03 |
25.18 |
0.025 |
|
High CRLF2 expression, logFC |
5.17 |
— |
0.0001 |
|
JAK2 mutations, % |
42 |
0 |
0.001 |
|
IKZF1 deletions, % |
61 |
24 |
0.001 |
|
RB1 deletions, % |
18 |
4 |
0.019 |
Conclusion
Taken together, the results suggest WTS can be used in routine diagnostics as a single assay, efficient stepwise approach to reliably stratify patients with B-ALL according to their individual genetic aberration profiles. Furthermore, the study found that WTS is superior to conventional detection methods in characterizing and quantifying gene expression profiles as well as identifying multiple transcriptional events in cases which lack entity-defining genetic abnormalities. However, standardized quality parameters and data analysis roadmaps are required to allow reproducibility and comparability amongst test centers.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

