All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme │ Diagnosis of Ph-like ALL
Do you know... Which of the following tests can identify probable Ph-like ALL in the shortest time from diagnostic sample receipt?
Philadelphia chromosome-like ALL (Ph-like ALL) is a high-risk subtype of precursor B-cell ALL (B-ALL) and accounts for 15–30% of B-ALL in older children, adolescents, and adults.
Our previous educational theme article was focused on the biology, genomic landscape, epidemiology, and prognosis of Ph-like ALL. In this educational theme article, we provide an overview of the different approaches available for the diagnosis of Ph-like ALL, summarized from articles by Harvey and Tasian,1 published in Blood Advances in 2020, and Frisch and Ofran, published in Haematologica in 2019.2
Ph-like ALL clinical diagnostics
Clinical diagnosis of patients with Ph-like ALL is challenging because of the associated heterogeneous genetic alterations (involving the 3′ kinase gene and 5′ fusion partner gene).1 There is no agreement on a standard approach for the diagnosis of patients who express a Ph-like gene signature, such patients usually present with a higher white blood cell count and are likely to remain minimal residual disease-positive following standard induction regimens.2 Several diagnostic strategies available for the identification of Ph-like ALL are described below.1
Gene expression analyses
Ph-like ALL was retrospectively identified based on extensive gene expression profiling. Of note, two large gene arrays using a 257-gene probe set and a 110-gene set shared only a minimal number of genes, with only the 257-gene probe set showing high expression of CRLF2 and JAK/STAT mutations. This discrepancy suggests that a definitive diagnosis should not rely only on the gene expression phenotype but rather on the identification of a genetic aberration in the cell signaling-related gene.2
Predictive analysis and low-density microarrays were the two predictors of the Ph-like ALL expression signature developed by researchers in the North America–based Children’s Oncology Group (COG). Both predictors use a continuous scale and apply cutoffs for determining the binary classification of Ph-like or not; however, the scores can also provide information about the probability of certain fusions.1
- Advantages: cost-effective; provides rapid results (within 48 hours).1
- Disadvantages: failed to detect mutations in about 15% of LDA-positive high-risk pediatric ALL patients.2
Cytogenetics and FISH analysis
Although cytogenetic analysis can identify major structural alterations, the majority of Ph-like ALL-associated alterations are cytogenetically ambiguous.1 Fluorescence in situ hybridization (FISH) panels are used for the primary screening of ALL. FISH probes can detect 3’ genes commonly involved in Ph-like ALL translocations, such as ABL1, ABL2, CRLF2, EPOR, JAK2, and PDGFRB. Abnormal 3’ gene results can be the first indicator of ABL class or CRLF2-R/JAK pathway-mutant Ph-like ALL, and provide the basis for further molecular characterization.1
- Advantage: provides rapid results (within 3 or 4 days)
- Disadvantage: often cannot identify the specific 5’ fusion gene partner
PCR
DNA-based polymerase chain reaction (PCR) assays have been useful in the detection of common Ph-like ALL-associated mutations, including ABL, JAK/EPOR, and CRLF2 translocations as well as IL7R indels.1,2
RT-PCR analyses
RNA/complementary DNA-based reverse-transcriptase polymerase chain reaction (RT-PCR) has been utilized for the molecular characterization of Ph-like ALL kinase fusions.1,2
- Advantages: provides rapid results (within 2 to 3 days); simultaneous testing of multiple kinase fusions.
- Disadvantages: significant potential for false-negative results; identification of kinase fusions with promiscuous breakpoints or previously unknown 5’ partners often not possible.
Flow cytometric immunophenotyping
Flow cytometry easily detects the increased surface thymic stromal lymphopoietin receptor (TSLPR; encoded by CRLF2) staining of ALL blasts, shown to be predictive of IGH-CRLF2 and P2RY8-CRLF2 rearrangements and CRLF2 F232 point mutations in primary Ph-like ALL cells.1 Genetic testing of TSLPR+ specimens should be used to further characterize the specific CRLF2 alterations as well as potential JAK and IL7R mutations.
- Advantage: cost-effective; provides rapid results (within 24 hours)
Next-generation sequencing platforms
Next-generation sequencing platforms provide comprehensive RNA and DNA-based hybrid capture or anchored multiplex PCR-based assays for Ph-like ALL characterization. Also, some platforms can identify novel fusions.1
- Advantages: cost-effective and widely available; capable of detecting more than 200–400 fusions/mutations in cancer-related genes.
- Disadvantage: slow turnaround for clinical results (2 to 4 weeks).
Whole-exome, transcriptome, and genome sequencing
Research-level transcriptomic/RNA sequencing is sometimes performed on specimens if kinase fusions, or other Ph-like mutations, are not identified in clearly low-density microarray positive ALL specimens.1
- Advantage: RNA sequencing is becoming widely available
- Disadvantage: high cost; slow turnaround for results
Based upon the above scientific and clinical advances, a practical, cost-effective, and time-efficient clinical algorithm to screen the Ph-like gene expression profile among all newly diagnosed patients with high-risk B-ALL was suggested by Harvey and Tasian (Figure 1).
Figure 1. Recommended clinical testing algorithm for the identification of patients with Ph-like ALL*
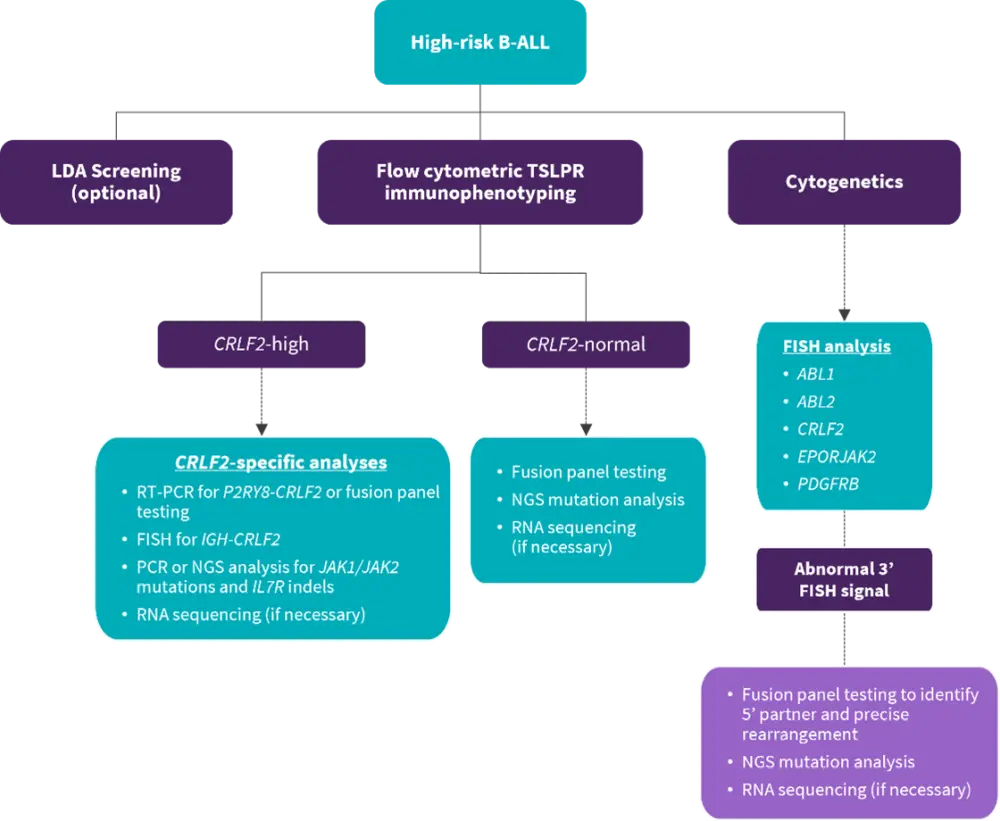
B-ALL, B-cell acute lymphoblastic leukemia; FISH, fluorescence in situ hybridization; LDA, low-density microarray; NGS, next-generation sequencing; PCR, polymerase chain reaction; RNA, ribonucleic acid; RT-PCR, reverse-transcriptase PCR; PDGFRB, platelet-derived growth factor receptor B; TSLPR, thymic stromal lymphopoietin receptor.
*Adapted from Harvey and Tasian1
Conclusion
The genetics of Ph-like ALL is heterogeneous and thus, diagnostically challenging. There is currently no consensus approach for the diagnosis and management of the disease. Overall, implementation of the suggested approach to screening high-risk patients with B-ALL for the Ph-like gene expression profile, as part of routine practice, could have a clinical benefit. The development of more sensitive and time-efficient diagnostic methods are needed to identify this high-risk patient population and will subsequently aid in finding appropriate treatment strategies.
“Look out for our following educational theme articles on the treatment of Ph-like ALL!”
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

