All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme│The genomic landscape, biology, epidemiology, and prognosis of Ph-like ALL
Do you know... Approximately 50% of patients with Ph-like ALL harbor rearrangements of which gene?
Our educational theme for this quarter is exploring the biology, genomic landscape, epidemiology, and prognosis of a relatively new subtype of acute lymphoblastic leukemia (ALL); Philadelphia chromosome (Ph)-like ALL. This educational theme will also include future articles on the diagnosis and treatment of Ph-like ALL.
Ph-like ALL is a high-risk subtype of B-cell precursor ALL, which is characterized by poor prognosis and outcomes; it harbors diverse genetic alterations (>70 discrete alterations), including a high rate of IKZF1 and other B-cell-associated transcription factor deletions, which activate cytokine receptor genes and kinase signaling pathways.1 Ph-like ALL occurs commonly in children, as well as adolescents and young adults (AYAs).2 Due to associated adverse outcomes, the World Health Organization classifies Ph-like ALL as a provisional leukemia subtype.3 The kinase-activated gene expression patterns of Ph-like ALL are very similar to BCR-ABL1-rearranged or Ph-positive (Ph+) ALL.3
Here, we summarize the biology, epidemiology, and prognosis of Ph-like ALL based on articles published by Tran and Tasian in Best Practice & Research Clinical Hematology in 2021,1 Tran and Loh Hematology Am Soc Hematol Educ Program in 2016,2 Harvey and Tasian in Blood Advances in 2019,3 and Iacobucci and Roberts in Genes in 2021.4
Biology and genomic landscape
The heterogenous genetic alterations in Ph-like ALL are categorized into four genomically-defined subsets based on the similarity of functional fusion partners and underlying kinase-activating lesions (Table 1)1:
- JAK/STAT pathway gene alterations;
- ABL class alterations;
- Ras pathway mutations; and
- rare kinase fusions.
Table 1. The landscape of Ph-like ALL kinase rearrangements, therapeutic targets, and clinical trials*
|
Ph-like ALL, Philadelphia chromosome-like acute lymphoblastic leukemia. |
|||
|
3’ kinase gene |
5’ fusion partner gene(s) |
Kinase inhibitors |
Clinical trial(s) |
|---|---|---|---|
|
JAK/STAT pathway alterations |
|||
|
CRLF2 |
CSF2RA, IGH, P2RY8 |
Ruxolitinib |
|
|
JAK2 |
ATF7IP, BCR, EBF1, ETV6, GOLGA5, HMBOX1, OFD1, PAX5, PCM1, PPFIBP1, RFX3, SMU1, SNX29, SSBP2, STRN3, TERF2, TPR, USP25, ZBTB46, ZNF274, ZNF340 |
Ruxolitinib |
|
|
EPOR |
IGH, IGK, IGL, LAIR1, THADA |
Ruxolitinib |
|
|
TSLP |
IQGAP2 |
Ruxolitinib |
— |
|
IL2RB |
MYH9 |
Ruxolitinib |
|
|
ABL class alterations |
|||
|
ABL1 |
CENPC, ETV6, FOXP1, LSM14A, NUP153, NUP214, RANBP2, RCSD1, SFPQ, SHIP1, SNX1, SNX2, SPTNA1, ZMIZ1 |
Dasatinib, imatinib, others |
|
|
ABL2 |
PAG1, RCSD1, ZC3HAV1 |
Dasatinib, imatinib |
|
|
CSF1R |
MEF2D, SSBP2, TBL1XR1 |
Dasatinib, imatinib |
|
|
PDGFRA |
FIP1L1 |
Dasatinib, imatinib |
|
|
PDGFRB |
ATF7IP, EBF1, ETV6, NUMA1, SNX29, SSBP2, TERF2, TNIP1, ZEB2, ZMYND8, ZNF608 |
Dasatinib, imatinib |
|
|
LYN |
GATAD2A, NCOR1 |
Dasatinib, imatinib |
— |
|
Other kinases |
|||
|
NTRK3 |
ETV6 |
Entrectinib Larotrectinib |
|
|
PTK2B |
KDM6A, STAG2, TMEM2 |
FAK inhibitors |
— |
|
FGFR1 |
BCR |
Ponatinib |
— |
|
FLT3 |
ZMYM2 |
FLT3 inhibitors |
— |
|
TYK2 |
MYB, SMARCA4, ZNF340 |
JAK1/3 inhibitor |
— |
|
BLNK |
DNTT |
— |
— |
|
CBL |
KANK1 |
— |
— |
|
DGKH |
ZFAND3 |
— |
— |
The most common alteration in Ph-like ALL is CRLF2 rearrangement, occurring in 64% of children and AYAs. 3 CRLF2 rearrangements may also occur at a lower frequency in children with standard risk (SR) B-ALL and in >50% of trisomy 21/Down syndrome (DS)-associated B-ALL; however, it is rare to discover other Ph‑like ALL-associated kinase fusions in patients with SR ALL.1
Deletions of IKZF1 and other lymphoid transcription factor genes are also seen in Ph+ ALL, resulting in aberrant marrow stromal adhesion, chemotherapy resistance, and inferior clinical outcomes.1 In addition, the new IKZF1plus molecular profile (defined as IKZF1 deletion co-occurring with one or more deletions in CDKN2A, CDKN2B, PAX5, or the pseudoautosomal region 1 [PAR1], and in the absence of ERG deletion) is associated with inferior outcomes, including a higher frequency of relapses, and could be used to refine the risk stratification criteria.1
Figure 1. Frequency of Ph-like ALL alterations in children, adolescents, and young adults*
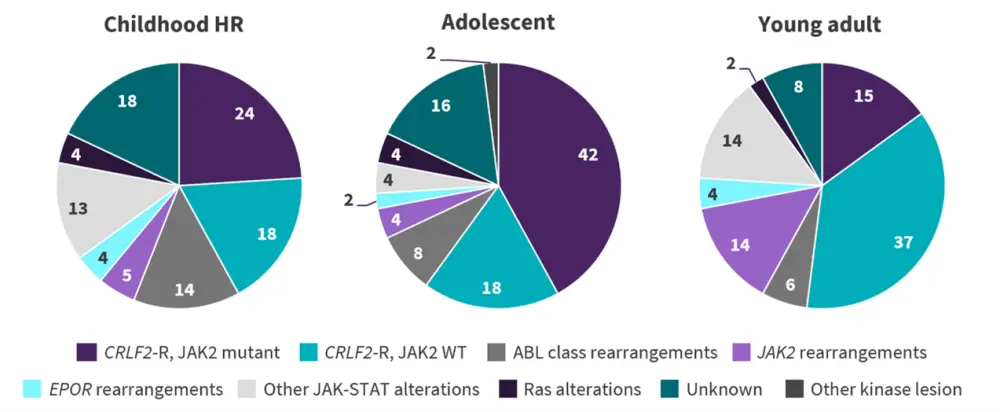
CRLF2-R, CRLF2-rearranged; HR, high risk; Ph-like ALL, Philadelphia chromosome-like acute lymphoblastic leukemia; WT, wild-type.
*Adapted from Tran and Loh.2
Key genomic alterations in Ph-like ALL are presented in Figure 2 and summarized below.
Figure 2. Key genomic alterations in Ph-like ALL*
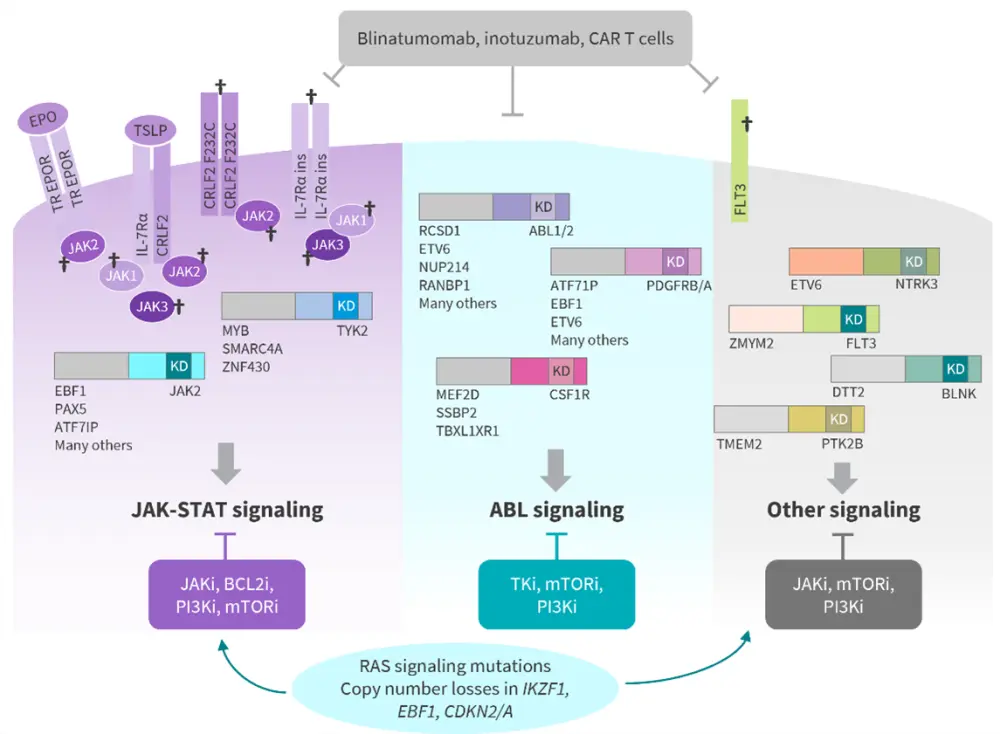
BCL2i, BCL2 inhibitors; CAR, chimeric antigen receptor; JAKi, JAK inhibitors; mTORi, mTOR inhibitors; Ph-like ALL, Philadelphia chromosome-like acute lymphoblastic leukemia; PI3Ki, PI3K inhibitors; TKi, tyrosine kinase inhibitors; TR, truncated.
*Adapted from Iacobucci I and Roberts KG.4
†Indicates the occurrence of a mutation.
JAK/STAT pathway gene alterations
Approximately 50% of patients with Ph-like ALL harbor CRLF2 rearrangements that occur either via focal deletion of PAR1 on chromosomes Xp22/Yp11, resulting in the P2RY8-CRLF2 fusion, or via translocation to the immunoglobulin heavy chain enhancer region on chromosome 14, resulting in IGH-CRLF2 rearrangement.1 P2RY8-CRLF2 fusion occurs more commonly in younger children with National Cancer Institute (NCI)-defined SR B-ALL and in those with DS-associated ALL.1 On the other hand, IGH-CRLF2 occurs mostly in AYAs, particularly in those of Hispanic/Latino or Native American ethnicity.1 CRFL2-rearranged (CRLF2-R) accounts for 24% of pediatric patients with NCI SR Ph-like ALL, 55% with high-risk disease, and 50−60% of AYA patients.4
Patients with Ph-like ALL, particularly those with CRLF2-R, have an increased risk of relapse due to the GATA3 single nucleotide polymorphism risk allele RS3824662.3 CRLF2-R is also significantly associated with mutations in JAK genes, most commonly the point mutation R683G in the JAK2 pseudokinase domain.1 Approximately 50−60% of children with DS-associated ALL have JAK2R683G mutations and CRLF2-R. JAK V658F, like the JAK2V617F mutation observed in adult patients with myeloproliferative neoplasms, occurs much less frequently in CRLF2-R Ph-like ALL.3 IL7R insertions and deletions occur in a small number of patients with CRLF2‑R that lack concomitant JAK1 or JAK2 mutations.3 Of note, 5−10% of patients with CRLF2-R ALL have distinctly different gene expression without the kinase-activated signature and therefore are not considered to have Ph-like disease.3
Mechanisms leading to activation of JAK/STAT signaling include EPOR rearrangements or kinase fusions involving JAK2, which release the normal auto-inhibition of the kinase domain.1 An increased frequency of EPOR rearrangements is observed with increasing age, with a peak in young adults. Sequence mutations and copy number variations in JAK1, JAK3, ILR7, SH2B3, IL2RB, and TYK2 are also associated with activation of the JAK/STAT pathway and occur more frequently in children compared with adolescents or young adults (14% vs 5% vs 7.3%, respectively). These cases often harbor chromosomal rearrangements expressing fusion oncoproteins involving transcription factor genes (EBF1, PAX5, and ETV6) and/or epigenetic regulators (CREBBP, SETD2, and ASXL1).1
ABL class alterations
ABL-class alterations account for ~10% of cases and are distinct from CRLF2-R and other JAK pathway-activating alterations.1 ABL-class alterations are more prevalent in children with NCI high-risk B-ALL compared with AYAs (17% vs 10%).1 Poor response to chemotherapy is observed in patients with ABL-class fusions, and the EBF1-PDGFRB fusion is associated with induction failure, indicating a potential mechanism for relapse.4 In EBF1-PDGFRB Ph‑like ALL, the T681I gatekeeper mutation has been demonstrated to be the most common resistance mutation to imatinib and dasatinib, with a trend towards increased risk of relapse in patients with T681I subclones.4
Ras pathway mutations
Ras pathway mutations are present in ~4% of Ph-like ALL cases, with mutations in the following Ras pathway genes: KRAS, NRAS, NF1, PTPN11, and CBL.1 These mutations are often subclonal, and commonly occur in hyperdiploid, hypodiploid, KTM2A-rearranged, and relapsed ALL. Ras pathway mutations may occur in conjunction with sentinel Ph-like translocations (e.g., ABL class, CRLF2, EPOR, or JAK2 fusions) or in isolation.1
Rare kinase fusions
Rare kinase fusions account for 5% of Ph-like ALL cases and involve NTRK3, BLNK, DGKH, FGFR1, PTK2B, FLT3, SH2B3, and TYK2. Among these cases, 1% harbor the ETV6-NTRK3 fusion4, which is reported to induce an aggressive ALL with in vitro and in vivo sensitivity to tyrosine receptor kinase inhibitors.1 As this fusion has been identified in a range of hematologic malignancies it is not unique to Ph-like ALL.1
Epidemiology and prognosis1
The prevalence of Ph-like ALL differs according to age, gender, range, ethnicity, and NCI-defined risk group. The pooled prevalence of Ph-like ALL is reported across the spectrum as 15.4%. Figure 3 illustrates the prevalence of Ph-like ALL by age and NCI risk group. Across all age groups, Ph-like ALL occurs more commonly in males than females, at a ratio of 1.5:1. Patients of Hispanic/Latino or Native American ethnicity, particularly those with CRLF2-R, are more predisposed to Ph-like ALL, at least in part due to the increased prevalence of the Ph-like ALL risk variant in GATA3 in this population.
In the pediatric population, the occurrence of Ph-like ALL is three times higher compared with Ph+ ALL.
Figure 3. Prevalence of Ph-like ALL*
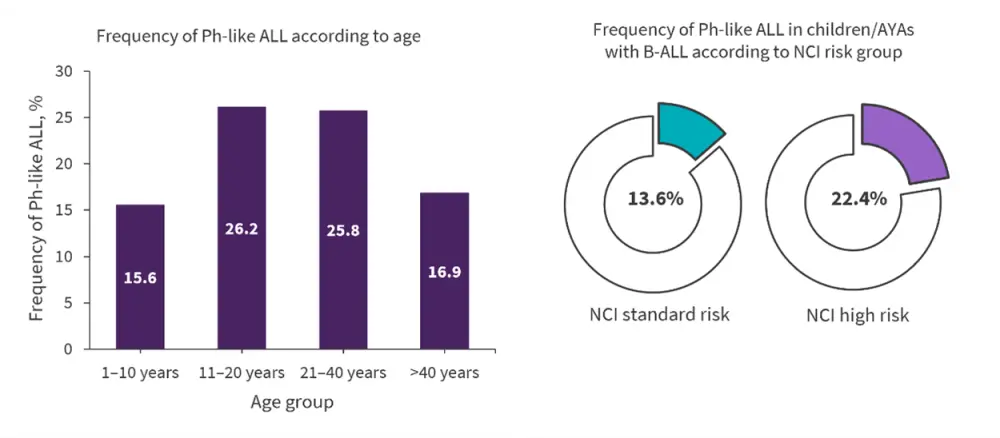
AYAs, adolescents and young adults; B-ALL, B-cell acute lymphoblastic leukemia; NCI, National Cancer Institute; Ph-ALL, Philadelphia chromosome-like acute lymphoblastic leukemia.
*Adapted from Tran and Tasian.1
Furthermore, patients with Ph-like ALL frequently exhibit adverse clinical features; significantly higher rates of hyperleukocytosis at diagnosis, end-induction minimal residual disease (MRD) positivity, and increased risk of treatment failure and relapse have been observed in patients with Ph-like ALL. Aggressive disease course and high rates of end-induction MRD or even induction failure are associated with patients harboring PDGFRB, JAK2, or EPOR fusions.
Compared with patients with Ph+ ALL, patients with Ph-like ALL have inferior survival outcomes across the age spectrum.
Conclusion
Ph-like ALL is emerging as a prevalent subtype of B-ALL, defined by its kinase-activated gene expression signature and associated genetic alterations. The prevalence of Ph-like ALL increases among children and AYAs, reaching a peak in young adults and is associated with a poor prognosis, with high rates of conventional treatment failure and relapse. As the genomic understanding of Ph-like ALL continues to develop, there is an opportunity to consider targeted therapeutic approaches that aim to reduce relapse rates and improve long-term survival for patients with Ph-like ALL across all ages.
“Look out for our following educational theme articles on the diagnosis and treatment of Ph-like ALL!”
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content

