All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit Know ALL.
The all Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the all Hub cannot guarantee the accuracy of translated content. The all and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The ALL Hub is an independent medical education platform, sponsored by Amgen, Autolus, Jazz Pharmaceuticals, and Pfizer and supported through an educational grant from the Hippocrate Conference Institute, an association of the Servier Group. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View ALL content recommended for you
Educational theme | Prognostic impact of chromosomal abnormalities and copy number alterations in B-ALL
Featured:
Genetic abnormalities have become well-established prognostic markers in adult acute lymphoblastic leukemia (ALL). The international UKALLXII/ECOG2993 trial identified one of the first widely used risk classifiers in adult ALL, showing that patients with BCR-ABL1, KMT2A-AFF1, low hypodiploidy/near triploidy (HoTr), or complex karyotype (CK) had a poor prognosis.1 The study also identified the presence of minimal residual disease (MRD) and age >40 years as risk factors for poor survival.
In the UKALL14 trial, a large cohort of adult patients with B-cell precursor ALL were treated based on risk assessment; patients with one or more of the above-mentioned risk factors were designated “high-risk” and assigned to allogeneic stem cell transplant (allo-SCT). In their study published in Leukemia, Anthony Moorman and colleagues,1 sought to identify the most relevant chromosomal abnormalities and copy number alterations (CNAs) in patients treated in the UKALL14 trial and proposed a revision to the UKALL genetic risk classification based on their results. In this ALL Hub educational theme article, we present a summary of the publication.
Study setup
A total of 652 patients with B-cell precursor ALL, aged between 25–65 years, were studied for the prognostic impact of chromosomal abnormalities and CNAs. Patients were assigned to high-risk treatment if they had any of the following: BCR-ABL1, KMT2A-AFF1, HoTr, CK, white blood cell count >30 × 109/L, MRD post phase 2 induction and age ≥41 years. High-risk patients were assigned to receive allo-SCT if they were fit and had an antigen-matched sibling or unrelated donor. Genetic analyses, including cytogenetics, FISH, MLPA, and SNP array, were performed on diagnostic bone marrow samples. The principal genetic abnormalities were BCR-ABL1, KMT2A-AFF1, other KMT2A fusions (together referred to as KMT2A-r), HoTr, CK (≥5 chromosomal abnormalities), JAK-STAT abnormalities (CRLF2 or JAK2 fusions), ABL-class fusions (ABL1, ABL2, PDGFRB, CSF1R fusions, except BCR-ABL1), ETV6-RUNX1, high hyperdiploidy (51–65 chromosomes), ZNF384 fusions (ZNF384-r), and t(1;9)(q21;p13)/TCF3-PBX1. Event- free survival, relapse rate and overall survival (OS) at 3 years were calculated by Kaplan–Meier methods. Hazard ratios were estimated using univariable and multivariable Cox regression models and p values <0.01 were considered statistically significant.
Findings
Frequency of genetic abnormalities
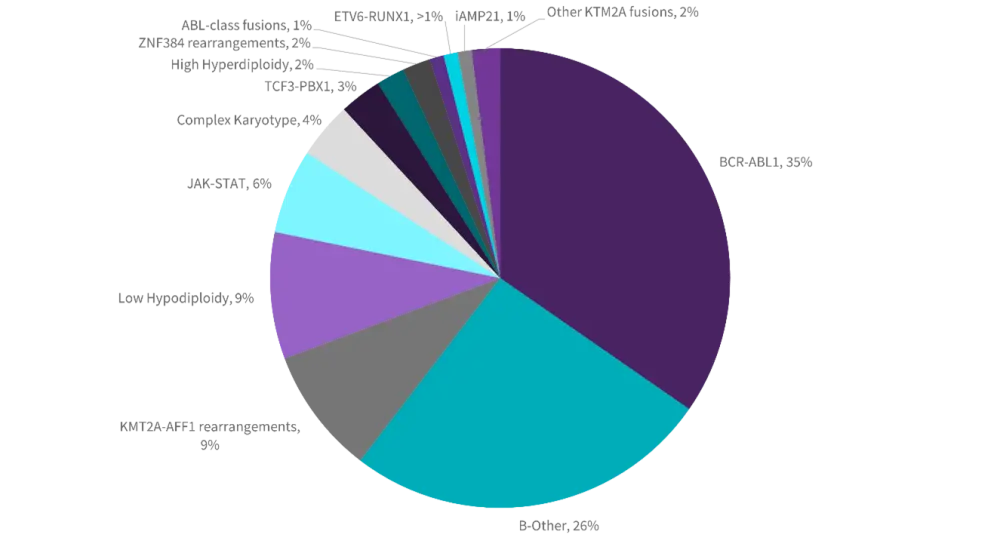
The median age of the study cohort was 46 years. Figure 1A shows the frequency of primary chromosomal abnormalities, the most frequent being BCR-ABL1, followed by KMT2A-r and HoTr. Chromosomal abnormalities associated with pediatric ALL were rare (four patients with iAMP21 and one patient with ETV6-RUNX1).
The frequency and co-occurrence of CNAs is shown in Figure 1B.
- Deletions of IKZF1 and CDKN2A/B were most frequent, occurring in 39% and 37% of patients, respectively.
- IKZF1 and CDKN2A/B were often co-deleted (p < 0.001) and over half of IKZF1 deletions fulfilled the criteria for IKZF1plus (IKZF1 plus PAX5, CDKN2A/B or PAR1 deletion).
- PAX5 and CDKN2A/B deletions also frequently co-existed (p < 0.001).
Figure 1. Frequency of primary chromosomal abnormalities (A) and co-occurrence of copy number alterations (B) in adults with B-cell precursor ALL
A.
B.
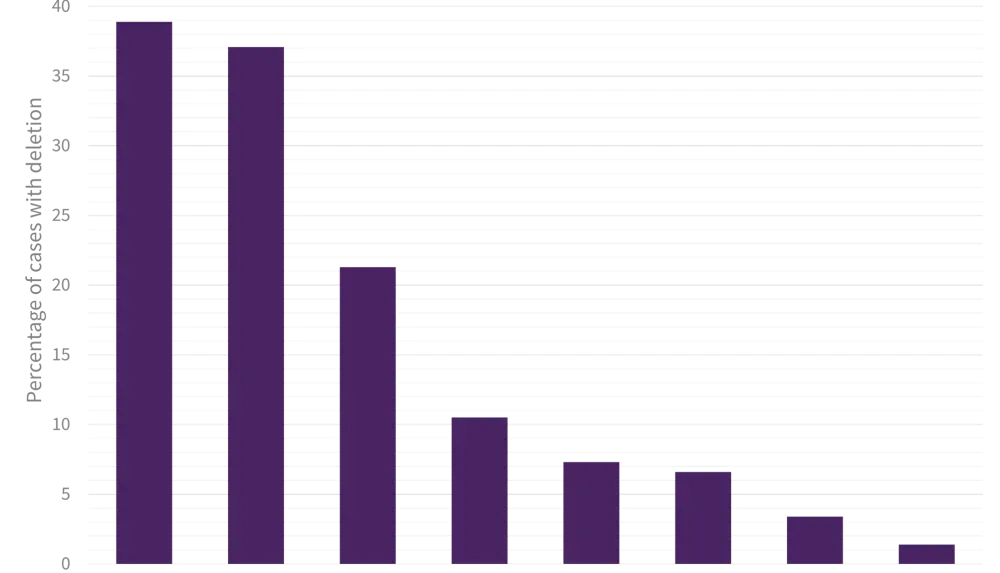
|
*Adapted from Moorman, et al. |
||||||||
|
Co-occurring deletions |
IKZF1 |
CDKN2A/B |
PAX5 |
BTG1 |
ETV6 |
RB1 |
EBF1 |
PAR1 |
|---|---|---|---|---|---|---|---|---|
|
IKZF1 |
170 |
|
|
|
|
|
|
|
|
CDKN2A/B |
82† |
162 |
|
|
|
|
|
|
|
PAX5 |
58 |
76 |
93 |
|
|
|
|
|
|
BTG1 |
37 |
30 |
21 |
46 |
|
|
|
|
|
ETV6 |
19 |
16 |
11 |
9 |
32 |
|
|
|
|
RB1 |
17 |
16 |
13 |
11 |
2 |
29 |
|
|
|
EBF1 |
14 |
2 |
4 |
4 |
2 |
3 |
15 |
|
|
PAR1 |
5 |
4 |
2 |
2 |
2 |
1 |
0 |
6 |
Relationship between genetic subtype and secondary abnormalities
There was a strong association between CNAs and genetic subtype.
- Patients with BCR-ABL1 had a higher frequency of IKZF1 deletions (60% vs 22%, p < 0.01) and IKZF1plus (32% vs 16%, p < 0.01) compared with other subtypes.
- Patients with KMT2A-r had few recurrent deletions and over half (56%) had no deletions.
- Patients with a JAK-STAT abnormality had a higher frequency of all CNAs, particularly IKZF1 deletions (63% vs 18%, p < 0.01) and IKZF1plus (54% vs 12%, p < 0.01).
- Patients with CK had a high frequency of PAX5 and/or CDKN2A/B and low frequency of IKZF1 deletions.
Outcome by genetic subtype and secondary abnormalities
Patients with HoTr or CK had a poor outcome despite being treated as high risk, with relapse and death rates double that of other patients.
- Relapse rates were 53% (hazard ratio [HR], 1.78; p = 0.018) and 60% (HR, 2.39; p = 0.005), respectively, compared with 32% for the overall cohort.
- The 3-year OS rates were 22% and 24%, respectively, compared with 54% for the overall cohort.
For patients classified as BCR-ABL1-like, those with an ABL-class fusion did not have a poor outcome, however those with a JAK-STAT abnormality, despite 83% of cases being treated as high risk, had high rates of MRD positivity at both timepoints (88% and 76%), a high relapse rate (56%) and poor OS (36%).
Relapse rates were higher but OS no worse for patients with KMT2A-AFF1 compared with other patients, and patients with BCR-ABL1 had similar relapse and OS rates to the remaining cohort. The outcome was also no worse for patients with TCF3-PBX1.
Overall, the CNAs detected in this study had no association with survival, although a biallelic CDKN2A/B deletion was associated with a poor outcome in the BCR-ABL1 cohort, with lower rates of event free survival (HR, 1.93; p = 0.01) and OS (HR, 1.88; p = 0.03) compared with those without the deletion.
Revised genetic risk classification
A revised genetic risk classification, comprising 4 groups with distinct genetics and/or outcomes was proposed:
- standard risk (SR): B-cell precursor ALL with ZNF384-r, HeH and other abnormalities
- high risk: KMT2A-r
- very high risk (VHR): HoTr, CK, JAK-STAT abnormalities
- tyrosine kinase activating fusions: BCR-ABL1, ABL-class fusions
Selected outcomes according to the revised risk groups are shown in Table 1. The 3-year OS of patients in the SR group was significantly higher than other patients (64% vs 47%; HR, 1.65; p < 0.001). Patients in the high-risk group had inferior outcomes compared with SR patients in univariate but not multivariate analysis, whereas patients in the VHR group had a worse prognosis than SR patients independent of gender, age, and white cell count.
Table 1. Outcome of UKALL14 patients according to the revised genetic risk classification*
|
*Adapted from Moorman, et al.1 |
||||||
|
Genetic risk group |
Frequency, % |
3-year relapse rate, % |
3-year EFS, % |
3-year OS, % |
Adjusted HR, p value for relapse rate† |
Adjusted HR, p value for EFS† |
|---|---|---|---|---|---|---|
|
Standard risk |
34 |
24 |
58 |
64 |
1 |
1 |
|
High risk |
10 |
51 |
37 |
45 |
1.49, 0.210 |
1.22, 0.409 |
|
Very high risk |
19 |
56 |
23 |
27 |
2.61, <0.001 |
2.34, <0.001 |
|
Tyrosine kinase activating fusions |
36 |
30 |
47 |
57 |
1.22, 0.304 |
1.35, 0.03 |
Conclusion
The prognostic impact of well-established chromosomal abnormalities alongside key copy number alterations was evaluated in this study of over 650 patients with B-cell precursor ALL treated on an MRD driven protocol. In addition to confirming the high-risk status of HoTr, KMT2A-r, and CK, this study found JAK-STAT abnormalities to also confer a poor outcome and proposed a revision to the genetic classification for adult ALL that will be of value for future studies.
References
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content


 Debalina Sarkar
Debalina Sarkar